Xie Wei’s group and Na Jie’s group report how histone modifications are transmitted from parents to their progeny in mammals
Research groups of Prof. Xie Wei and Prof. Na Jie at Tsinghua University have published a research paper in Nature on September 15th, entitled “Allelic reprogramming of the histone modification H3K4me3 in early mammalian development”. Xie Wei’s group has also published a paper in Molecular Cell on September 16th, entitled “Resetting Epigenetic Memory by Reprogramming of Histone Modifications in Mammals”. These two papers focuse on epigenetic reprogramming in mouse early development, together revealing how histone modifications are transmitted from parents to progeny in mammals.
Histone modifications are fundamental epigenetic regulators that control many crucial cellular processes. However, whether these marks can be passed on from gametes to the next generation in mammals is a long-standing question that remains unanswered. To study the reprogramming of histone modifications in early development, the Xie lab developed a low-input ChIP–seq method by combining an improved ChIP protocol that reduces sample loss and a highly sensitive library preparation method (TELP). This method was therefore termed STAR ChIP–seq (small-scale TELP-assisted rapid ChIP–seq). By applying this method to histone modifications H3K4me3 and H3K27me3 at a panel of mouse developmental stages, the authors provide a panoramic view of the landscapes of histone marks on each parental genome from developing gametes to post-implantation embryos.
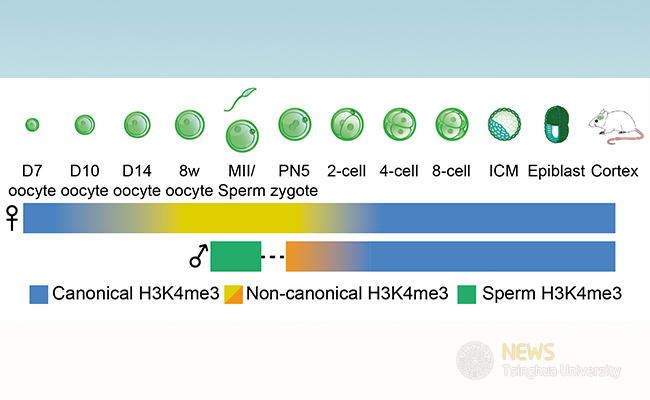
In the work described in Nature, the authors developed STAR ChIP–seq and reported how a key histone mark H3K4me3 was transmitted from gametes to the next generation. Upon fertilization, extensive reprogramming occurs on the paternal genome, as sperm H3K4me3 peaks are rarely observed in zygotes, suggesting a global erasure of paternal H3K4me3. Paternal H3K4me3 peaks are then readily observed after major zygotic genome activation at the late two-cell stage. By contrast, H3K4me3 from oocytes is inherited after fertilization which persists until the 2-cell stage before zygotic genome activation (ZGA) starts. Unexpectedly, the authors find a surprising non-canonical form of H3K4me3 (ncH3K4me3) in full-grown and mature oocytes, which exists as broad peaks at promoters and a large number of distal loci. Such broad H3K4me3 peaks are in contrast to the typical sharp H3K4me3 peaks restricted to CpG-rich regions of promoters. Notably, ncH3K4me3 in oocytes overlaps almost exclusively with partially methylated DNA domains. It is then inherited in pre- implantation embryos, before being erased in the late two-cell embryos, when canonical H3K4me3 starts to be established. The removal of ncH3K4me3 requires zygotic transcription but is independent of DNA replication-mediated passive dilution. Finally, the authors showed that downregulation of H3K4me3 in full-grown oocytes by overexpression of the H3K4me3 demethylase KDM5B is associated with defects in genome silencing, indicating ncH3K4me3 may function as a repressive mark, instead of an active mark, in oocytes and early stage embryos.
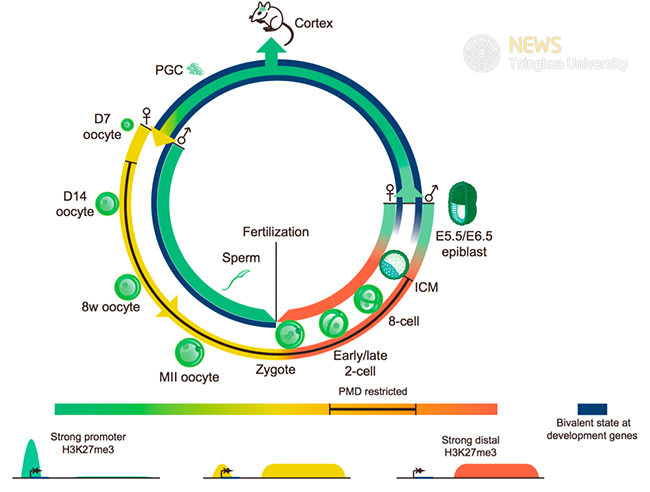
In a separate study reported in Molecular Cell, the authors focused on another crucial histone modification - H3K27me3. Polycomb group proteins and the related histone modification H3K27me3 can maintain the silencing of key developmental regulators and provide cellular memory. H3K27me3 usually appears at the promoters of key developmental genes. It remains unclear if such epigenetic memory can be passed on to the progeny. Similar to H3K4me3, the authors found sperm H3K27me3 is globally erased after fertilization. On the contrary, oocyte H3K4me3 appears to be selectively retained in zygotes. The authors showed that upon fertilization, the promoter H3K27me3 peaks at Hox and other developmental genes are extensively lost while distal H3K27me3 peaks are inherited. The resulting allele-specific H3K27me3 patterns persist to blastocysts before being converted to canonical forms in postimplantation embryos. As a result, both H3K4me3/H3K27me3 bivalent promoter marks are largely erased during preimplantation development and are readily restored at developmental genes only after implantation. Together, these data revealed widespread resetting of epigenetic memory and striking plasticity of epigenome during gametogenesis and early development.
For the work reported in Nature, Dr. Xie Wei from School of Life Sciences at Tsinghua University and Dr. Na Jie from School of Medicine at Tsinghua University are the co-corresponding authors. PhD students Bingjie Zhang, Hui Zheng (CLS program) from School of Life Sciences at Tsinghua University, Bo Huang (CLS program) from Academy for Advanced Interdisciplinary Studies of Peking University, and Wenzhi Li from School of Medicine, Tsinghua University are the co-first authors. Collaborators include Dr. Feng Xu’s group at Singapore Institute for Clinical Sciences, A-STAR Institute in Singapore, Dr. Shaorong Gao’s group at School of Life Sciences and Technology at Tongji University, Dr. Anming Meng’s group from School of Life science at Tsinghua University, and Dr. Kehkooi Kee’s group from School of Medicine at Tsinghua University. For the study published in Molecular Cell, Dr. Xie Wei is the corresponding author. PhD students Hui Zheng, Bo Huang, and Bingjie Zhang are the co-first authors. Collaborators include Dr. Feng Xu’s group at Singapore Institute for Clinical Sciences, A-STAR Institute in Singapore. These two studies are supported by the animal facility, the sequencing facility, and the Biocomputing facility at Tsinghua University. This work is supported by the funding provided by the National Basic Research Program of China (973 program), the National Natural Science Foundation of China, the National Basic Research Program of China, the funding from the THU-PKU Center for Life Sciences and the Youth Thousand Scholar Program of China.
Paper link:
http://www.nature.com/nature/journal/vaop/ncurrent/full/nature19361.html
http://www.cell.com/molecular-cell/fulltext/S1097-2765(16)30479-8
(edited by Li Huashan)

