Maojun Yang’s Group Published the Structure of Respirasome on Cell
On December 1st, Professor Yang’s group from the School of Life Science, Tsinghua University published an article entitled Structure of mammalian respiratory supercomplex I1III2IV1 on Cell, reporting for the first time the atomic resolution cryo-EM structure of mammalian respirasome (1). The current work is a breakthrough by Prof. Yang in this area, after reporting the medium-high resolution structure of respirasome (2) in Nature on September 21st, 2016.
On December 2nd, Professor Yang’s group held a news conference in Rom Building presenting their important results. Honored guests included Guowei Sang, Zihe Rao, Linfang Wang, Haidong Wu, Zhe Yang, Shuxia Gu, Dong Liu, Yijun Qi, Xinquan Wang and Hongwei Wang (Figure 1).

Figure 1. Honored guests
In the latest paper, through optimizing the protein purification procedure and data processing method, Professor Yang’s group successfully pushed the resolution to 3.6 angstrom which subsequently revealed the detailed interaction between subunits in CI, identified the linking subunits between CI, CIII and CIV, discovered the critical role of lipid molecules in the structure of respirasome, and presented a new electron transfer model. This achievement enables us to deeply understand the organization of mammalian respiratory chain and the mechanism of proton pumping and electron transfer, and provides critical information in treating cellular-respiration related diseases.
Respiration is one of the most fundamental biological process in all living organisms. Dysfunction of mitochondrial respiratory chain complexes generates reactive oxygen or nitrogen species, and this type of dysfunction is implicated in many human diseases1–3, including Alzheimer’s and Parkinson’s diseases, multiple sclerosis, Friedreich’s ataxia and amyotrophic lateral sclerosis. As such the respiratory chain is always a topic in structural biology research. Structural study of respiratory supercomplexes could be traced back to the year of 2005, when Dudkina determined the first structure of SCI1III2 purified from Arabidopsis thaliana at a resolution of 18 angstrom. Then in 2007, Jesco provided a structural model of SCIII2IV1-2 from yeast mitochondria at a resolution of 15 angstrom. Several years later, two groups independently reported the structure of SCI1III2IV1 from bovine heart at the resolution of 22 angstrom and 19 angstrom respectively in 2011. Finally Professor Yang’s group was able to push the resolution to 3.6 angstrom. (Figure 2)
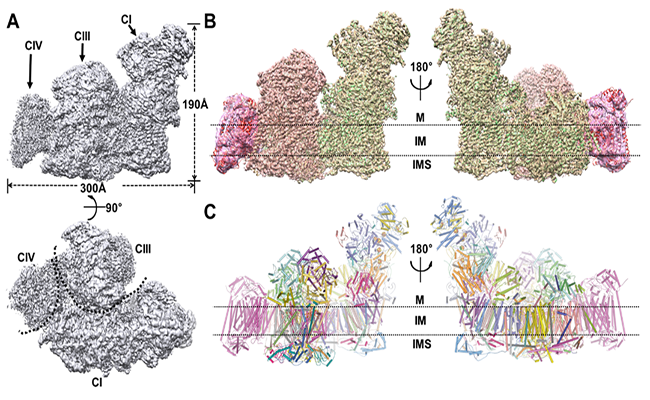
Figure 2. Overall structure of respirasome
Based on the structure, Professor Yang provided a new electron transfer model. The UQH2 is released from the CI and diffuses to the Qi site close to the heme bH of CIII, then transfers one electron to heme bH and changes to semiQ (SQ) radical. After that, the heme bH transfers the electron to heme bL, meanwhile accepts another electron from the SQ radical and the oxidized UQ would be released from the Qi site. After both of the heme bLs acquired an electron and the ISP proteins changed to the “b” state, the electrons could be transferred to the two [2Fe-2S] clusters, and then the ISP proteins change to the “c1” state to transfer the electrons to the heme c1 from their [2Fe-2S] clusters. Finally, each heme c1 transfers the single electron to Cyt.c. Together with the transfer of electrons, the two protons dissociated from the UQH2 might be translocated from the Qi site to the IMS using the redox energy released by electron transfer. In conclusion, in each cycle of electron transfer cascade one molecule of UQH2 distributes two electrons to two separate electron acceptors, Cyt.c, and in the meanwhile CIII translocates two protons from UQH2 to IMS. (Figure 3)
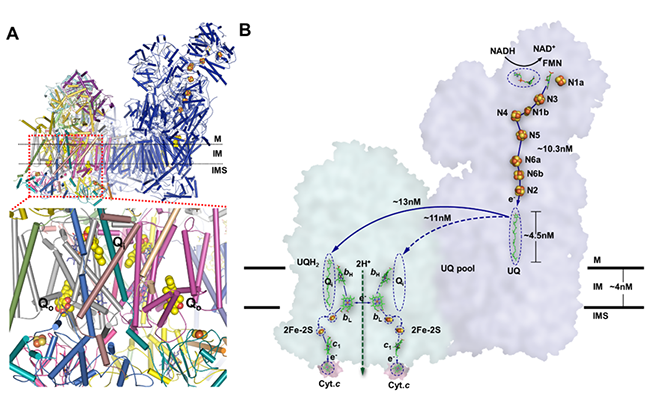
Figure 3. Electron transfer pathway in CI and CIII
Ph.D student Meng Wu and Jinke Gu, Master student Runyu Guo and Yushen Huang from Prof. Yang’s group are co-first authors of this work. Professor Maojun Yang is the corresponding author. Crosslink mass spectrometry was performed by Dan Tan from the National Institution of Biological Science. Data collection was performed on the Cyro-EM Platform, and computing work was supported by Tsinghua University High Performance Computing Cluster. This work was founded by Beijing Advanced Innovation Center for Structural Biology, Tsinghua University Branch of China National Center for Protein Sciences, Tsinghua-Peking Joint Center for Life Sciences, Ministry of Education Key Laboratory of Protein Science, Ministry of Science and Technology, National Outstanding Young Scholar Science Foundation, and National Natural Science Foundation of China.

