Yigong Shi’s Group published the EM structure of a yeast step II catalytically activated spliceosome on Science, yielding a near-complete mechanistic picture on the splicing cycle
On December 16, 2016, Professor Yigong Shi’s group in School of Life Sciences, Tsinghua University, published a Research Article entitled “Structure of a Yeast Step II Catalytically Activated Spliceosome” in Science. The paper reported the cryo-EM structure of Saccharomyces cerevisiae spliceosome at the state just before the second step of splicing reaction. This structure illustrates the mechanism of spliceosome remodeling after finishing the first step of splicing reaction and further reveals the molecular mechanism of pre-mRNA splicing.
In eukaryotes, because of the existence of introns, the pre-mRNA transcribed from DNA cannot be translated into proteins directly. The introns must be removed by RNA splicing and the resulting mature mRNA can form the template of protein synthesis on ribosome. The removal of introns need two sequential transesterification reactions: First, the phosphodiester bond at the 5’ splice site (SS) is attacked by the 2’-hydroxyl of an adenosine of the branch point sequence (BPS) in the intron, which generates a free 5’ exon and an intron lariat-3’ exon. Subsequently, the 3’-hydroxyl of the 5’ exon attacks the phosphodiester bond at the 3’SS, leading to exon ligation and excision of the lariat intron. Through these two steps, introns of different number and length from pre-mRNA are removed and the rest of the exons are ligated in a specific order (Fig 1).
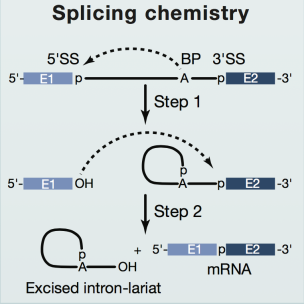
Figure 1 The two steps of RNA splicing(Cell)
RNA splicing is accomplished by a dynamic mega-complex known as spliceosome. To catalyze splicing of the pre-mRNA, spliceosome undergoes extremely precise stepwise assembly, subsequently forming a series of spliceosomal complexes. According to the reaction state and biochemical characterization, these complexes can be defined as B, Bact , B*, C, C*, P, and ILS complexes. It is one of most challenging biological projects to capture the different activation and catalysis state of spliceosome and determine their structures.
In August 2015, Yigong Shi’s group made the first breakthrough, reporting the high resolution cryo-EM structure of Schizosaccharomyces pombe (S.pombe) intron lariat spliceosome (ILS complex) at 3.6 angstrom, the first near-atomic structure of the spliceosome in the world. In July 22, Yigong Shi’s group published two back to back Science Research Articles, reporting the near-atomic resolution cryo-EM structures of Saccharomyces cerevisiae activated spliceosome (Bact complex) and catalytic step I spliceosome (C complex),completely display the conformation of pre-mRNA substrate and catalytic snRNAs, as well as the assembly of spliceosomal protein components before and after the first step transesterification reaction. However, no high resolution structure was available to show the details of spliceosome catalyzing the second splicing reaction.
In their recent Science article, Shi and coworkers acquired good samples of S.cerevisiae spliceosome and reported the three-dimensional structure of step II catalytically activated spliceosome (C* complex) at the overall resolution of 4.0 angstrom by single particle cryo-EM. This structure further provides important information of RNA splicing, describing the composition and conformational changes of spliceosome, especially the catalytic center between the first and second transesterification reaction. The local resolution for the core region is at 3.5 angstrom and provided insights into the second transesterification reaction.
Since August, 2015, Shi’s group reported five high resolution structures of the crucial spliceosomal complexes, that is, the pre-assembled U4/U6.U5 tri-snRNP at 3.8 angstrom, the activated spliceosome Bact complex at 3.5 angstrom, the catalytic step I spliceosome C complex at 3.4 angstrom, the step II catalytically activated spliceosome C* complex at 4.0 angstrom and the intron lariat spliceosome ILS complex at 3.6 angstrom. These five high resolution structures almost cover all the working states of spliceosome assembly and catalysis, providing extremely abundant information which subsequently aids the development of the RNA splicing field.
Prof. Yigong Shi is the corresponding author. Dr. Chuangye Yan (post doc from School of Life Sciences and Center for Life Sciences), Ruixue Wan (PhD student from School of Medicine) and Rui Bai (PhD student from School of Life Sciences) are the co-first authors of this paper. Gaoxingyu Huang (PhD student from School of Life Sciences) participates in this study. EM images were acquired at the Tsinghua University Branch of China National Center for Protein Sciences (Beijing). Data processing was performed on the “Explorer 100” cluster system of Tsinghua National Laboratory for Information Science and Technology, the Computing Platform of China National Center for Protein Sciences (Beijing), and Mr. Donghui Wang (Chairman of United Electronics). This research was funded by the Beijing Innovation Center for Structural Biology and the National Natural Science Foundation of China.
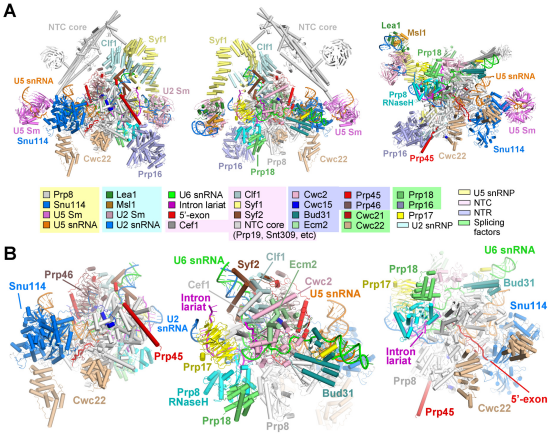
Figure 2 the structure of C* complex
The original link:
http://science.sciencemag.org/content/early/2016/12/14/science.aak9979.full
Related publications:
http://science.sciencemag.org/content/early/2016/01/06/science.aad6466
http://science.sciencemag.org/content/early/2015/08/19/science.aac8159
http://science.sciencemag.org/content/early/2015/08/19/science.aac7629
http://science.sciencemag.org/content/early/2016/07/20/science.aag0291
http://science.sciencemag.org/content/early/2016/07/20/science.aag2235

