Junbiao Dai’s group published an article in Science, reporting the design and synthesis of the budding yeast chromosome XII
The research group of Dr. Junbiao Dai from School of Life Sciences at Tsinghua University published an article in Science on March 10th, 2017, entitled “Engineering the ribosomal DNA in a megabase synthetic chromosome”. This is one of several findings of a package of seven papers published as the cover story for Science. This work reported the design and synthesis of the longest eukaryotic chromosome (synthetic chromosome 12 or synXII) in history, with length of 976,067bp. The synthetic chromosome was designed based on its native counterpart and assembled by a two-step method developed in the lab, which eventually generated a yeast strain with normal biological function. The researchers also engineered the highly repetitive ribosomal RNA coding sequences (rDNA) residing in ChrXII. In addition, Dr. Junbiao Dai’s group also participated in the synthesis of chromosome VI (with New York University Langone Medical School, USA), chromosome II (with BGI and University of Edinburgh, UK) and chromosome V (with Tianjin University), and analyzed the 3D structure of the synthetic chromosomes (with Institute Pasteur).
To understand the origin of life, scientists have long been challenged whether an artificial living cell could be produced in the lab from scratch. The development of gene synthesis technology has made it possible to generate the entire genetic materials of an organism and the genome of viruses, phages and bacteria have been redesigned and synthesized. Most important of all, the synthetic genomes are capable of replication and produce progenies once incorporated into a cell. In the past few years, a group of researchers formed an international consortium to re-design and synthesize the 12 Mb yeast genome – a project named Sc2.0. It aims to construct a “designer” genome after introducing changes based on an arbitrary set of principles such as eliminating the repetitive sequences and transposons, and incorporating thousands of LoxP sites for future genome manipulation. By applying the central principle of synthetic biology, “build to understand”, this project would gain deeper understanding of the yeast genome to answer many basic biological questions and to make the budding yeast a more efficient system for industrial application.
Among the sixteen chromosomes, Chromosome XII is unique as one of the longest yeast chromosomes (approximately 1 million base pairs) and additionally encodes the highly repetitive ribosomal DNA locus, which forms the well-organized nucleolus. A 976,067-base pair linear synXII was designed based on the native chromosome XII and chemically synthesized. synXII was assembled using a two-step method (Figure 1), involving successive megachunk integration to produce six semi-synthetic strains, followed by meiotic recombination-mediated assembly, yielding a full-length functional chromosome in S. cerevisiae.
The ribosomal gene cluster (rDNA) on synXII was left intact during the assembly process and subsequently replaced by a modified rDNA unit. The same synthetic rDNA unit was used to regenerate rDNA at three distinct chromosomal locations. The rDNA signature sequences of the internal transcribed spacer (ITS), used to determine species identity by standard DNA barcoding procedures, were swapped to generate a Saccharomyces synXII strain that would be identified as Saccharomyces bayanus. Remarkably, these dramatic DNA changes had no detectable phenotypic consequences under various laboratory conditions.
Synthesis of synXII highlights our ability to design and construct megabase-sized chromosomes and to manipulate the highly repetitive rDNA, laying the foundation to rewrite ultra large and complex genome in the future. It also speaks to the remarkable overall flexibility of the yeast genome.
Prof. Junbiao Dai is the corresponding author. Weimin Zhang (PhD student from Center for Life Sciences), Guanghou Zhao and Zhouqing Luo (PhD students from the Peking University-Tsinghua University-NIBS (PTN) Joint Graduate Program) are the co-first authors of this paper. Prof. Guo-qiang Chen and Prof. Qingyu Wu from Tsinghua University, Prof. Joel S. Bader from Johns Hopkins University, USA, Prof. Yizhi Cai from University of Edinburgh, UK, Prof. Jef D. Boeke from NYU Langone Medical School, USA and Mr. Jiwei Lin from China-based industrial partner Wuxi Qinglan Biotechnology Inc. contributed to this work. DNA and RNA sequencing services were provided by Jianhuo Fang from sequencing facility at Tsinghua University. This research was funded by the National Natural Science Foundation of China, the National Basic Research Program of China (973 Program), Ph.D. Programs Foundation of Ministry of Education of China, Research Fund for the Doctoral Program of Higher Education of China, the Tsinghua University Initiative and startup fund from Youth “thousand talent” program.
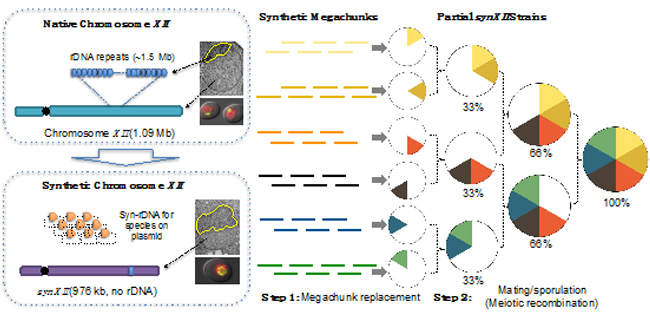
Fig. 1 Hierarchical assembly and subsequent restructuring of synXII
http://science.sciencemag.org/content/sci/355/6329/eaaf3981.full.pdf

